The genomes of hundreds of types of cancer have been sequenced by various teams around the world. The sequencing information is being used to target cancer therapies for maximum patient benefit. Studies show that patients who receive targeted biologics have improved survival. Clinicians now perform multigene sequencing for cancer patients and prescribe therapies specifically adapted for each individual patient's case.
Genomics tools are currently available to clinicians. You can find "genetically informed cancer care" at My Cancer Genome. You can find the Integrated Genomic Database for NSCLC (IGDB.NSCLC) here.
These improved outcomes appear to be accompanied by significant additional cost. According to a report at thepharmaletter.com, the addition of 11 novel therapies to the NSCLC pharmacopeia will boost the retail value of this market to nearly $13 billion by 2024.
Integrated Genomic Database of Non Small Cell Lung Cancer
Shao-Hsuan Kao et al published an integrated genomic database of non small cell lung cancer (NSCLC) called IGDB.NSCLC. The research was conducted by Dr. Kao and colleagues at the Institute of Biochemistry, Microbiology and Immunology, Chung Shan Medical University, Clinical Laboratory, Chung Shan Medical University Hospital in Taiwan, and was published in Nucleic Acids Research.
As far as we know, IGDB.NSCLC is the largest integration of lung cancer genomic resources providing multiple levels of evidence to search for the concordantly altered targets and to prioritize putative NSCLC genes for future studies. In addition to the database with various searching options and user-friendly interfaces, we provided concordant somatic alterations based on the genome-wide CNAs data with co-localization of altered gene expression, aberrant miRNA expression, somatic mutation and genes in association with clinicopathological features.
The high concordance of CNA data with these somatic alterations and clinical features in IGDB.NSCLC suggested the quality of data integration with experimental validations, the heterogeneity of genetic pathways to NSCLC and the important roles of genomic alterations involved in tumor formation of NSCLC.
The future development of integrating other NSCLC resources into IGDB.NSCLC will focus on increasing the data of clinicopathological features for dissecting the altered genes involved in tumor progression and on somatic alteration data from next-generation sequencing results. In conclusion, IGDB.NSCLC is an invaluable resource for selecting putative cancer genes in NSCLC to better understand the heterogeneous tumorigenic mechanisms and for developing useful strategies in clinical applications to prolong the life of lung cancer patients.
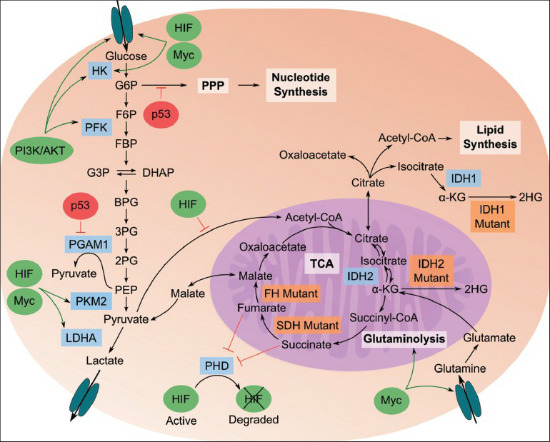
in cancerous cells.
Molecular Evidence Development Consortium is Funded
According to PR Newswire, the non-profit Molecular Evidence Development Consortium (MED-C) has signed or agreed to sign funding agreements with the following companies:
- Roche/Genentech
- Celgene
- Eli Lilly and Company
- Illumina Inc.
- Thermo Fisher Scientific
With this arrangement, MED-C has acquired the majority of the funds it need to launch its first project, a registry of lung cancer sequencing data.
“Although the complete MED-C NGS [next generation sequencing] registry will encompass many different disease states, the starting point is the 'N1N Registry' where patients with newly diagnosed advanced non-small cell lung cancer will have the opportunity to have their tumor tested by high-quality, standardized NGS, be treated by the latest standard of care and have de-identified clinical outcomes collected in a centralized, open-access, database,” Dane J. Dickson, MD told PR Newswire.
“This project focuses the efforts and strengths of all major stakeholders (patients, providers, payors, regulators, pharma, and industry/laboratories) into building one of the greatest advances in personalized medicine ever undertaken. The generous support of these companies in both donations and expertise has allowed us to get this started. We hope that many others will join with them in helping us rapidly advance medicine in a way that we can only accomplish by working together.” Dr. Dickson is the Chief Operating Officer of MED-C.
Comprehensive Genomic Profiles of Small Cell Lung Cancer
A similar joint initiative has assembled a comprehensive genomic profiles of murine small cell lung cancer. This vast multicenter trial was overseen by Julie George and Jing Shan Lim, both of the Department of Translational Genomics at the Center of Integrated Oncology Cologne–Bonn, Medical Faculty, at the University of Cologne, Germany. The report was published in Nature.
We have sequenced the genomes of 110 small cell lung cancers (SCLC), one of the deadliest human cancers. In nearly all the tumors analyzed we found bi-allelic inactivation of TP53 and RB1, sometimes by complex genomic rearrangements. Two tumors with wild-type RB1 had evidence of chromothripsis leading to overexpression of cyclin D1 (encoded by the CCND1 gene), revealing an alternative mechanism of Rb1 deregulation. Thus, loss of the tumor suppressors TP53 and RB1 is obligatory in SCLC. We discovered somatic genomic rearrangements of TP73 that create an oncogenic version of this gene, TP73Δex2/3. In rare cases, SCLC tumors exhibited kinase gene mutations, providing a possible therapeutic opportunity for individual patients. Finally, we observed inactivating mutations in NOTCH family genes in 25% of human SCLC. Accordingly, activation of Notch signaling in a pre-clinical SCLC mouse model strikingly reduced the number of tumors and extended the survival of the mutant mice.
